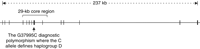Microcephalin, a Gene Regulating Brain Size, Continues to Evolve Adaptively in Humans
- Patrick D. Evans1,2,
- Sandra L. Gilbert1,
- Nitzan Mekel-Bobrov1,2,
- Eric J. Vallender1,2,
- Jeffrey R. Anderson1,
- Leila M. Vaez-Azizi1,
- Sarah A. Tishkoff4,
- Richard R. Hudson3,
- Bruce T. Lahn1,*
+ Author Affiliations
- ↵* To whom correspondence should be addressed. E-mail: blahn@bsd.uchicago.edu
Abstract
The gene Microcephalin (MCPH1) regulates brain size and has evolved under strong positive selection in the human evolutionary lineage. We show that one genetic variant of Microcephalin in modern humans, which arose ∼37,000 years ago, increased in frequency too rapidly to be compatible with neutral drift. This indicates that it has spread under strong positive selection, although the exact nature of the selection is unknown. The finding that an important brain gene has continued to evolve adaptively in anatomically modern humans suggests the ongoing evolutionary plasticity of the human brain. It also makes Microcephalin an attractive candidate locus for studying the genetics of human variation in brain-related phenotypes.
The most distinct trait of Homo sapiens is the exceptional size and complexity of the brain (1, 2). Several recent studies have linked specific genes to the evolution of the human brain (3–12). One of these is Microcephalin (7, 8); mutations in this gene cause primary microcephaly [MCPH; Online Mendelian Inheritance in Man (OMIM) accession 251200] (13, 14). MCPH is defined clinically as severe reductions in brain size coupled with mental retardation, but remarkably, an overall retention of normal brain structure and a lack of overt abnormalities outside of the nervous system (15–17). This led to the notion that the brains of MCPH patients function normally for their size and that genes underlying MCPH are specific developmental regulators of brain size (15–17).
Microcephalin is one of six known loci, named MCPH1 through MCPH6, for which recessive mutations lead to MCPH (14, 18–23). For four of these, the underlying genes have been identified as Microcephalin (MCPH1), CDK5RAP2 (MCPH3), ASPM (MCPH5), and CENPJ (MCPH6) (14, 21, 23). Patients with loss-of-function mutations in Microcephalin have cranial capacities about 4 SD below the mean at birth. As adults, their typical brain size is around 400 cm3 (whereas the normal range is 1200 to 1600 cm3), and the cerebral cortex is especially small (13, 14). Microcephalin is suggested to control the proliferation and/or differentiation of neuroblasts during neurogenesis. This postulate was consistent with several observations. First, mouse Microcephalin is expressed prominently in the proliferative zones of the embryonic brain (14). Second, the Microcephalin protein contains several copies of the BRCT domain that is found in cell cycle regulators, such as BRCA1 (14, 24). Finally, cell culture studies indeed suggested a role of Microcephalin in regulating cell cycle (25–27).
The finding that Microcephalin is a critical regulator of brain size spurred the hypothesis that it might have played a role in brain evolution (16, 28). Consistent with this hypothesis, phylogenetic analysis of Microcephalin revealed signatures of strong positive selection in the lineage leading to humans (7, 8). Here, we examine the possibility that positive selection has continued to operate on this gene after the emergence of anatomically modern humans.
The human Microcephalin locus has 14 exons spanning about 236 kb on chromosome 8p23 (14) (Fig. 1). We previously sequenced all the exons in 27 humans (8). When re-analyzing the data, we noticed that one haplotype had a much higher frequency than the other haplotypes. Additionally, this haplotype differed consistently from the others at position 37995 of the genomic sequence (counting from the start codon) or position 940 of the open reading frame. This polymorphism falls in exon 8 and changes amino acid residue 314 from an ancestral aspartate to a histidine. (This polymorphism is described as G37995C with G denoting the ancestral allele.)
Genomic structure of the human Microcephalin gene. The region sequenced in the 89-individual Coriell panel is bracketed.
To investigate whether positive selection has acted on the high-frequency haplotype, we resequenced 23.4 kb of a 29-kb region centered around the G37995C polymorphism (Fig. 1). Sequencing was performed on a panel of 89 individuals from the Coriell Institute, which broadly represents human diversity (see SOM). To assign the ancestral state of polymorphisms, we also sequenced the common chimpanzee. Several GC-rich segments were not sequenced because of technical difficulties. The resulting sequence data contained 220 polymorphic sites, including 213 single-nucleotide polymorphisms (SNPs) and 7 insertion/deletion polymorphisms (indels) (table S1).
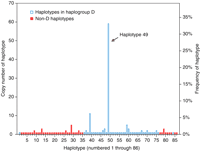
Haplotypes were inferred using the PHASE 2.1 program (29, 30). A total of 86 haplotypes were identified along with their frequencies (Fig. 2 and table S2). One haplotype, denoted 49, had a much higher frequency than the other haplotypes. It had the derived C allele at the G37995C SNP site and corresponded to the high-frequency haplotype in the aforementioned exon-only polymorphism survey (8). In the Coriell panel, haplotype 49 had a frequency of 33% (59 out of 178 chromosomes) and is found in all the populations sampled in the panel. The remaining 85 haplotypes varied in frequency from 0.6 to 6.2% (1 to 11 chromosomes).
Frequencies of 86 inferred Microcephalin haplotypes in the 89-individual Coriell panel. Haplotypes in haplogroup D are indicated by blue-edged bars; non-D haplotypes are indicated by solid red bars.
Positive selection on an allele can increase the frequency of the haplotype bearing the allele while maintaining extended linkage disequilibrium (LD) around that allele (31–36). Our data on haplotype 49 are consistent with these signatures of selection. We formally tested the statistical significance of positive selection using the previously established coalescent model (37, 38). Given the slight uncertainty in haplotype inference, we considered only the 18 individuals in the Coriell panel who are homozygous for haplotype 49 (table S1).
By simulation, we calculated the probability of obtaining 18 or more individuals (out of 89) who are homozygous for a single haplotype across a region of 220 segregating sites under neutral evolution. Here, recombination and gene conversion rates were set to values previously established for the Microcephalin locus (39), and a demographic model with a severe bottleneck followed by exponential growth was assumed (see SOM). Prior studies have shown that the bottleneck specified here is likely to be much more stringent than that associated with the real demographic history of human populations (40, 41); thus, the test is conservative (38). Under these parameters, the probability of obtaining 18 homozygotes out of 89 is highly significant (P = 0 based on 5,000,000 replicates).
We then tested several additional demographic models, including (i) constant size, (ii) very ancient expansion, (iii) very recent expansion, (iv) repeated severe bottlenecks with subsequent expansion, and (v) population structure with between two and five subpopulations (see SOM). All produced exceedingly significant results. Even though the exact demographic history of humans is yet to be defined, our tests are highly significant under a broad range of demographic scenarios, which furthers the argument that the statistical significance is unlikely to be altered by reasonable variations in the supposed human demography. We also tested the significance of the inferred haplotype data (i.e., the significance of having 59 copies of haplotype 49 among 178 chromosomes), which similarly produced highly significant results. These data strongly suggest that haplotype 49 was driven to high frequency by positive selection. However, our data do not address whether the positive selection is frequency-dependent selection, heterozygote advantage, or simple additive positive selection.
Using the G37995C polymorphism as a diagnostic site, we divided all the haplotypes into two groups: those that carry the derived C allele and those that carry the ancestral G allele. We designated the former group as haplogroup D (where D stands for “derived”). It includes 43 haplotypes that together have a 70% frequency in the Coriell panel, and haplotype 49 is the predominant member (table S2). Although the derived C allele at the G37995C site only provides an operational definition for haplogroup D, several observations make evident that haplogroup D is systematically different from the non-D haplotypes. First, this haplogroup consists exclusively of haplotype 49 or its minor variants, whereas non-D haplotypes show much greater sequence divergence from haplogroup D chromosomes. This greater divergence is because haplogroup D and non-D haplotypes have multiple fixed differences relative to each other in addition to G37995C (table S2). The only exceptions are a few recombinant haplotypes between D and non-D chromosomes (discussed below). Second, for sites that are polymorphic within haplogroup D chromosomes (excluding recombinants between D and non-D chromosomes), the non-D chromosomes are invariably monomorphic for the ancestral alleles. These data indicate that haplogroup D constitutes a genealogical clade of closely related haplotypes that is altogether separate from the more distantly related non-D haplotypes (again, excluding recombinants between D and non-D chromosomes, which represent mixed genealogies).
Collectively, the above observations support an evolutionary scenario with two aspects. First, haplotype 49 swept from a single copy to high frequency in a short period of time. Second, during the sweep, minor variants of haplotype 49 emerged through rare mutations and recombinations. These variants, together with haplotype 49, make up haplogroup D. Haplotype 49 evidently represents the most recent common ancestor (MRCA) of haplogroup D, because it consistently has the ancestral allele for the sites polymorphic within haplogroup D.
We next estimated the coalescence age (i.e., time to MRCA) of haplogroup D chromosomes in the Coriell panel. We used the average number of mutations from the MRCA of a haplogroup clade to its descendant lineages as a molecular clock for estimating the age of the clade (42, 43). This approach is known to be unbiased by demographic history (42). The age of haplogroup D was found to be ∼37,000 years, with a 95% confidence interval of 14,000 to 60,000 years. In comparison, the coalescence age of all the chromosomes in the Coriell panel is about 1,700,000 years. The emergence of anatomically modern humans has been estimated to be 200,000 years before present (44). Haplogroup D is obviously much younger, which indicates that positive selection was at work in a period considerably postdating the emergence of anatomically modern humans in Africa. We note that the age of haplogroup D coincides with the introduction of anatomically modern humans into Europe about 40,000 years ago, as well as the dramatic shift in the archeological record indicative of modern human behavior, such as art and the use of symbolism (i.e., the “Upper Paleolithic revolution”) (45).
If haplogroup D indeed experienced a recent selective sweep, it should show low polymorphism and an excess of rare alleles (46). To confirm this, we calculated nucleotide diversity (π) and Tajima's D for the 47 individuals who are homozygous for haplogroup D chromosomes, and we compared these values to those of the non-D chromosomes. The π value of the D chromosomes is lower, by a factor of 12, than that of the non-D chromosomes (0.000077 and 0.00092, respectively), even though the D chromosomes represent about 70% of the chromosomes in the panel. Tajima's D, which is a summary statistic for the frequency spectrum of alleles, is –2.3 for haplogroup D (whereas it is –1.2 for the non-D chromosomes). This strongly negative Tajima's D indicates a starlike genealogy for haplogroup D chromosomes (47). Thus, both summary statistics contrast sharply between D and non-D chromosomes and are consistent with the recent age and rapid expansion of haplogroup D. We note that these calculations do not provide a statistically stringent test of positive selection, because they are done on subsets of the genealogy. Nevertheless, they do reveal qualitative signatures of positive selection that further corroborate the more stringent statistical tests described earlier.
Another sign of a positive selective sweep is extended LD around the selected allele. This is apparent in the region of Microcephalin investigated here, where haplogroup D chromosomes show near-complete LD across the entire region. The only exceptions are haplotypes 1, 68, and 84 (each found in a single copy in the Coriell panel), which are recombinants between D and non-D chromosomes as evidenced by recombination tracts (table S2). The remaining 121 copies of haplogroup D chromosomes show no evidence of recombination. By comparison, the non-D chromosomes do not display any significant LD across the region.
To probe the extent of LD beyond the 29-kb core region, we sequenced the Coriell panel for two segments of about 3 kb each, situated at the beginning and end of the gene separated from each other by about 235 kb. In these flanking regions, there is clear evidence of LD decay from the core region, which supports the idea that selection has most likely operated on a site (or sites) around the core region. Our present data cannot resolve the exact site(s) of selection, and the G37995C nonsynonymous SNP used to define haplogroup D is just a candidate.
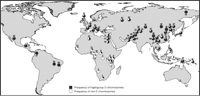
To obtain a more detailed frequency distribution of haplogroup D across the globe, we analyzed a much larger human population panel containing 1184 globally diverse individuals. We genotyped the diagnostic G37995C SNP in this panel to infer the frequency of haplogroup D chromosomes (Fig. 3). Geographic variation was observed, with sub-Saharan populations generally having lower frequencies than others. The statistic for genetic differentiation, FST, is 0.48 between sub-Saharans and others, which indicates strong differentiation (48) and is significantly higher than the genome average of 0.12 (P < 0.03 based on previously established genomewide FST distribution) (49). Such population differentiation may reflect a Eurasian origin of haplogroup D, local adaptation, and/or demographic factors such a bottleneck associated with human migration out of Africa 50,000 to 100,000 years ago.
Global frequencies of Microcephalin haplogroup D chromosomes (defined as having the derived C allele at the G37995C diagnostic SNP) in a panel of 1184 individuals. For each population, the country of origin, number of individuals sampled, and frequency of haplogroup D chromosomes are given (in parentheses) as follows: 1, Southeastern and Southwestern Bantu (South Africa, 8, 31.3%); 2, San (Namibia, 7, 7.1%); 3, Mbuti Pygmy (Democratic Republic of Congo, 15, 3.3%); 4, Masai (Tanzania, 27, 29.6%); 5, Sandawe (Tanzania, 32, 39.1%); 6, Burunge (Tanzania, 28, 30.4%); 7, Turu (Tanzania, 23, 15.2%); 8, Northeastern Bantu (Kenya, 12, 25%); 9, Biaka Pygmy (Central African Republic, 32, 26.6%); 10, Zime (Cameroon, 23, 8.7%); 11, Bakola Pygmy (Cameroon, 24, 10.4%); 12, Bamoun (Cameroon, 28, 17.9%); 13, Yoruba (Nigeria, 25, 24%); 14, Mandenka (Senegal, 24, 16.7%); 15, Mozabite [Algeria (Mzab region), 29, 53.5%]; 16, Druze [Israel (Carmel region), 44, 60.2%]; 17, Palestinian [Israel (Central), 40, 63.8%]; 18, Bedouin [Israel (Negev region), 44, 54.6%]; 19, Hazara (Pakistan, 20, 85%); 20, Balochi (Pakistan, 23, 78.3%); 21, Pathan (Pakistan, 23, 76.1%); 22, Burusho (Pakistan, 25, 66%); 23, Makrani (Pakistan, 24, 62.5%); 24, Brahui (Pakistan, 25, 78%); 25, Kalash (Pakistan, 24, 62.5%); 26, Sindhi (Pakistan, 25, 78%); 27, Hezhen (China, 9, 77.8%); 28, Mongola (China, 10, 100%); 29, Daur (China, 10, 85%); 30, Orogen (China, 10, 100%); 31, Miaozu (China, 9, 77.8%); 32, Yizu (China, 10, 85%); 33, Tujia (China, 10, 75%); 34, Han (China, 41, 82.9%); 35, Xibo (China, 9, 83.3%); 36, Uygur (China, 10, 90%); 37, Dai (China, 9, 55.6%); 38, Lahu (China, 10, 85%); 39, She (China, 9, 88.9%); 40, Naxi (China, 10, 95%); 41, Tu (China, 10, 75%); 42, Cambodian (Cambodia, 11, 72.7%); 43, Japanese (Japan, 27, 77.8%); 44, Yakut [Russia (Siberia region), 25, 98%]; 45, Papuan (New Guinea, 17, 91.2%); 46, NAN Melanesian (Bougainville, 18, 72.2%); 47, French Basque (France, 24, 83.3%); 48, French (France, 28, 78.6%); 49, Sardinian (Italy, 26, 90.4%); 50, North Italian [Italy (Bergamo region), 13, 76.9%]; 51, Tuscan (Italy, 8, 87.5%); 52, Orcadian (Orkney Islands, 16, 81.3%); 53, Russian (Russia, 24, 79.2%); 54, Adygei [Russia (Caucasus region), 15, 63.3%]; 55, Karitiana (Brazil, 21, 100%); 56, Surui (Brazil, 20, 100%); 57, Colombian (Colombia, 11, 100%); 58, Pima (Mexico, 25, 92%); 59, Maya (Mexico, 25, 92%).
Previous studies have shown that Microcephalin is a specific regulator of brain size (13, 14) and that this gene has evolved under strong positive selection in the primate lineage leading to Homo sapiens (7, 8). Here, we present compelling evidence that Microcephalin has continued its trend of adaptive evolution beyond the emergence of anatomically modern humans. The specific function of Microcephalin in brain development makes it likely that selection has operated on the brain. Yet, it remains formally possible that an unrecognized function of Microcephalin outside of the brain is actually the substrate of selection. If selection indeed acted on a brain-related phenotype, there could be several possibilities, including brain size, cognition, personality, motor control, or susceptibility to neurological and/or psychiatric diseases. We hypothesize that D and non-D haplotypes have different effects on the proliferation of neural progenitor cells, which in turn leads to different phenotypic outcomes of the brain visible to selection.
Supporting Online Material
www.sciencemag.org/cgi/content/full/309/5741/1717/DC1
Materials and Methods
Tables S1 and S2
References and Notes
- Received for publication 18 April 2005.
- Accepted for publication 14 June 2005.